Project Description
now browsing by category
BioWES Testing
IMARES – Ijmuiden, The Netherlands
BioWES system was used to develop protocols in “Growth of juvenile yellowtail kingfish (Seriola lalandi) at different temperature“ experiment. Contact person: Abbing Wout. http://www.wageningenur.nl/nl/Expertises-Dienstverlening/Onderzoeksinstituten/imares.htm
NOFIMA – Sunndalsora, Norway
BioWES system was used to study the growth and survival at different experimental unit scales. Contact person: Asa Maria O. Espmark. http://www.nofima.no/en/artikkel/4869733954035201089
The Institute of Microbiology, Czech Academy of Sciences, Třeboň, Czech Republic
Development and testing of the BioWES LC-MS module. Contact person: Petra Kučerová. http://www.alga.cz/en
ULPGC – Grupo de Investigación en Acuicultura, Spain
BioWES system was used to study the tag effect on growth and mortality of fish in fingerlings of 300 mg. Contact person: Juan Manuel Afonso Lopez. www.ulpgc.es
WU – Wageningen, The Netherlands
BioWes system was used for development of protocol for the „Effect of dietary oxygen demand on feed intake in fish“ experiment. Contact person: Ep Edding. http://www.wageningenur.nl/en/Expertise-Services/Chair-groups/Animal-Sciences/Aquaculture-and-Fisheries.htm
Data sharing and management – web solution
Metadata central storage
The central data storage will be realized as combination of local data storage (located on the institution) for raw data and one central data storage selected meta-data. Central data storage will be based on distributed relation ORACLE database to deal with the amount and structure of the meta-data. The data structures in central database will be defined generally to cover all the different meta-data types and to upgrade the structures in the future.
Data availability
The central data storage will serve as the first option for searching the experimental data through meta-data and allow the user to find the proper experiment and results. All the meta-data will be available using the XLM data structures exchange.
The solution based on relational database allows:
– standard access to the metadata using SQL standard language and support of wide range of database interfaces
– high accessibility of the system will be allowed by distribution to more computers using GRID technology of CLUSTERING
– fast information search – the usage of data structures indexing and advanced algorithms of query execution plans will minimize the time response of the data storage
– standardized system control – the commercial database ensure the safe and secured operation of the database
Information system of central data storage:
The central database will be in cooperation with the information system. The information system will provide the parameterization of the central storage, user accounts control and policy. The data from central database will be presented using the web presentations to the different kind of the scientific and laic public controlled by the access rights.
Central storage interface:
The interface will be used as an interpreter between central data storage, local data storage and visualization framework.
Visualization framework:
The user friendliness of the central database will be supported by the visualization framework. The visualization framework will be implemented as several software modules and interface for extension of information system. The visualization framework will allow simple and intelligible visualization and comparison of the meta-data and result of searches. It will be based on the mix of existing modules and third party modules. The third party modules could be plugged into the central data storage for the user of the system. The modules will be focused on raw data processing and data mining and aggregation of the meta-data. Standard interface of the central data storage will allow the user to upload the mined information back into the central storage, describe it and integrate it into current structures.
One part of the web solution will be the offer of tools for raw data post-processing. These tools will be highly specialized tools for experimental data processing. The tools can be used by anyone to produce the metadata from raw experimental data and share the metadata using BioWes web solution. The list of the tools can be extended by any third party tool for biological data processing. Several tools will be developed directly under the project:
– Cell time lapse image processing and representation
– LC-MS measurements filtration and analysis
– Software for behavior analysis of aquatic organisms
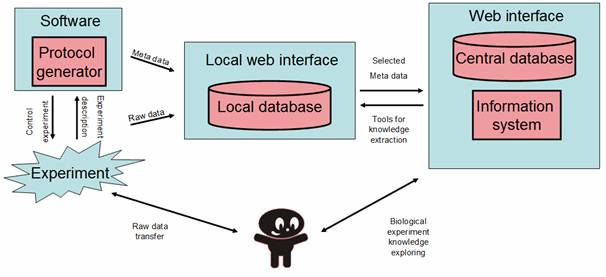
Local data management tools
The main purpose of local data management tool is to organize and store the raw experimental data directly on the site of the institution (experimenter). The local data management tool will provide the functionality of data storing, searching, filtration and reporting. The tool will be connected to the Protocol generator to support the reporting of experiments on the higher level of metadata. Local data management will be realized as a specialized database that will be optimized to the type of experimental data produced by particular institution. The database with uniform interface will be modified according to the needs of the particular experimenter to reach the aims of different experiments.
The local management tool will provide the communication module (interface) to global data management tool (web based data sharing). The global data management tool will be used for meta-data sharing between different institutions and the public. The process of communication between local and global data management tools will be under full control of the institution. Therefore only the meta-data can be shared with the rest of scientific community or the public. The advantage of this approach is the direct control of „what I am sharing with the others“.
There are many different solutions and technologies for raw data storage among the institutions. The review of technologies will be realized during the first year of the project to select the best technology for local data management tool.
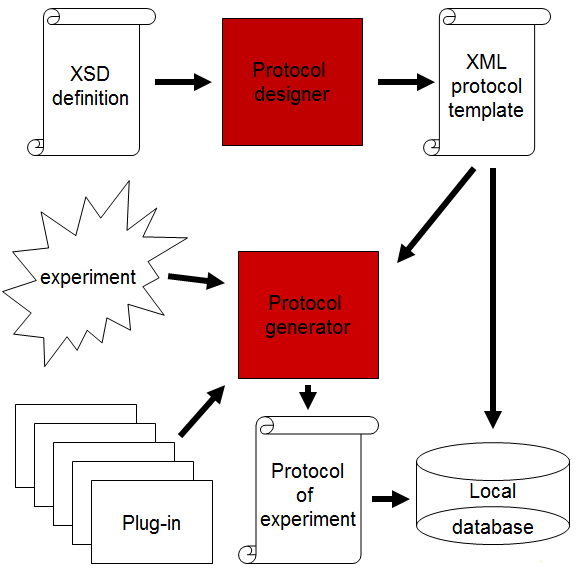
Protocol generator
It is an independent application that should ensure the repeatability and correctness of the biological experiments. The tool is designed to lead the experimenter through the particular type of experiment as a supervisor and help. Protocol generator has two purposes: the first one is to check that the procedure of the experiment has been done precisely and the second one is to produce all important settings that are part of the experiment in the form of report on the experiment. The method to ensure precise realization of experiment is to check if all necessary parameters and steps of the experiment have been set and done. The list of necessary parameters and steps for the particular type of experiment comes up from the analysis of biological experiment for different research institutes.
The description of the experiment can be created by the user for specific experiment. Graphical user interface Protocol designer has been implemented for this purpose. The protocol template is created by the person who fully understands the experiment and who can define all the important conditions of the experiment. The user can use 10 basic components for definition of the protocol. The protocol of experiment can be shared among the people who realize the experiment (students) to ensure the repeatability of the experiment.
The protocol is internally defined by XML language and has to follow the XSD scheme prescribed for experiment description. Templates of protocols can be stored in the local database for future usage. The template can be later modified for new experiment to speed up the process.
Main advantage of the electronic protocol is that there is a direct link between the protocol and experimental data. Both are stored in the central database and can be used for obtaining future data.
Protocol generator supports external plugins for mining information about setting of devices from external files. The plugins can read the information about some parameters of experiment form files produced by measurement device (magnification of microscope) and fill it into the protocol. Plugins are using open interface and therefore it can be created by the users for specific devices.
Collection and analysis of experimental procedures
The first year of the project is focused on monitoring and analysis of the situation on the field of biology experiments. Many research institutes and organizations will participate on this most important part of understanding the needs of the experimentalists. The needs of different types of research organizations on biology experiments will be collected and analyzed. The questionnaire with questions concerning the type of experiment, needed equipment, experimental procedure, data storage, data sharing etc. has been prepared for this purpose. The output of the first year analysis will be knowledge about data types produced by biology experiments, realization of experiments and data management. Each of the participants of the initial analysis will be able to influence the final solution of the project. They can influence the design and functionality of the Protocol generator, local data management tool and global web based data sharing tools. It is a unique opportunity to get free solution for the whole chain of experimentation actions.
BioWES – Project description
Project BioWes is a collaborative project of University of South Bohemia in České Budějovice represented by Institute of Complex Systems and private company dataPartner ltd. that works on delivery and implementation of information and control technologies. The type of the solution of the project is applied research. Institute of Complex Systems is responsible for the research part of the project focused on biology experiments (see more here). DataPartner is going to deliver database and web based solutions for data management and sharing (see more here).
The project BioWes is inspired by several similar projects that try to solve a substantial contemporary problem of sharing big amount of experimental data. There are several projects that offer the solution for data sharing (for different types of data). The problem is that the amount of data produced by experimentalist is constantly increasing and the speed of internet will always be a step behind. The only way how to share experimental data on between researchers is to share the metadata. Metadata means the overall knowledge about the experiment that consist of detailed knowledge of experimental procedure and knowledge that can be mined from the data automatically or manually by post-processing. The data itself is meaningless without any additional knowledge concerning the experiment. There is no project that can offer experimental data sharing based on the sharing of knowledge.
The main reason why to share the experimental data is to save money and time necessary for experimentation and to compare the results between different experimenters. Data sharing and especially metadata sharing can be understood as the advertisement of the experiments of a particular experimenter. Experimental data sharing and comparison can help to improve experimental procedures and definition of standards in this area.
Project BioWes is focused on the area of biology experiments. These experiments produce one of the biggest amounts of data and impose the highest requirements on the preciseness of experimentation to reproduce the experiment.

 D5 Creation
D5 Creation